Sometimes, it is just fun to do some simple math and get nice figure, why not drawing some bifurcation diagrams.
How it works
To draw a bifurcation diagram, we need to find some recurrence relation that exhibits chaotic behaviour. A nice place to find them is to use Euler method for specific dynamic system such as logistic systems.
For this specific recurrence:
![]()
Where ![]() is our parameter and
is our parameter and ![]() will be some initial condition, there are multiple different behaviour (we may want to discard some few first points) for the series: stability, multiple equilibrium points and chaos.
will be some initial condition, there are multiple different behaviour (we may want to discard some few first points) for the series: stability, multiple equilibrium points and chaos.
We can implement the recurrence function as follow:
def model(x, r):
return r * x * (1. - x)And we create a function that compute the ![]() first terms and may keep only the
first terms and may keep only the ![]() last terms:
last terms:
def diagram(r, x=0.1, n=1200, m=200):
if not isinstance(x, np.ndarray):
x = np.full(r.size, x)
xs = []
for i in range(n):
if i >= n - m:
xs.append(x)
x = model(x, r)
return np.array(xs).TNow we can compute the 30 first terms of the recurrence relation for several values of parameter ![]() to see what are the behaviour of the generated series:
to see what are the behaviour of the generated series:
rtest = np.array([0., 0.5, 1.5, 3.0, 3.1, 3.5, 4.0])
xtest = diagram(rtest, n=30, m=30)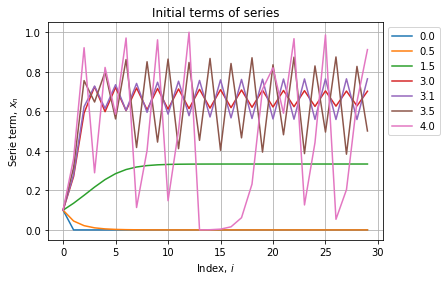
As expected all series starts at 0.1, some converges to a constant other oscillate between two or more specific values. We can see further to check if the behaviour stabilizes:
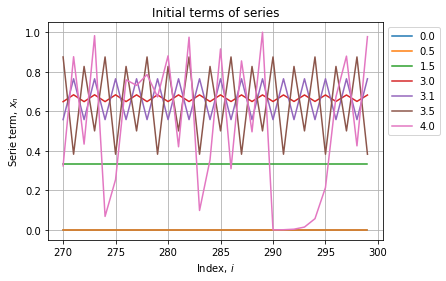
Now instead of plotting last series terms wrt to their index, we will assess them wrt the parameter ![]() . We also will test multiple values of initial conditions
. We also will test multiple values of initial conditions ![]() and assign them different colors.
and assign them different colors.
rlin = np.linspace(-2, 4, 500)
xlin = np.linspace(-0.1, 0.1, 2)
clin = np.linspace(0., 1., xlin.size)
colors = plt.get_cmap("cool")(clin)The bifurcation diagram will then we plotted as follow:
fig, axe = plt.subplots(figsize=(8, 6))
for x0, color in zip(xlin, colors):
x = diagram(rlin, x=x0, n=600, m=100)
axe.plot(rlin, x, ',', color=color)
axe.set_title("Bifurcation diagram")
axe.set_xlabel("Parameter,  ")
axe.set_ylabel("Serie term,
")
axe.set_ylabel("Serie term,  ")
axe.grid()
")
axe.grid()And renders as:
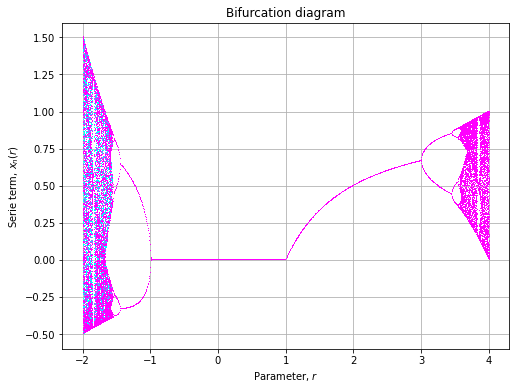
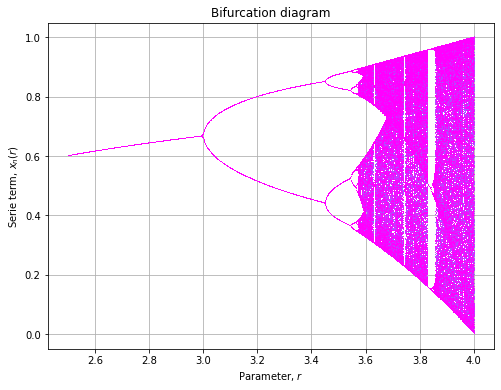
Examples
Lets plot more of them with this all on one plotting function:
def plot(model, rlin=None, xlin=None, colors=None, n=1200, m=200, rmin=-20, rmax=20, rsize=1000, cmap="cool", axe=None):
if rlin is None:
rlin = np.linspace(rmin, rmax, rsize)
if xlin is None:
xlin = np.linspace(-0.5, 0.5, 10)
if colors is None:
clin = np.linspace(0., 1., xlin.size)
colors = plt.get_cmap(cmap)(clin)
if axe is None:
fig, axe = plt.subplots(figsize=(8, 6))
for x0, color in zip(xlin, colors):
x = diagram(model, rlin, x=x0, n=600, m=100)
axe.plot(rlin, x, ',', color=color)
axe.set_title("Bifurcation diagram")
axe.set_xlabel("Parameter,  ")
axe.set_ylabel("Serie term,
")
axe.set_ylabel("Serie term, 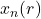 ")
axe.grid()
return axe
")
axe.grid()
return axeBiological system
![]()
def sigmoid(x, r):
return 5.821 * np.tanh(1.487 * x) - r * np.tanh(0.2223 * x)
axe = plot(
model=sigmoid,
rmin=5, rmax=30,
xlin=np.array([-0.1, 0.1])
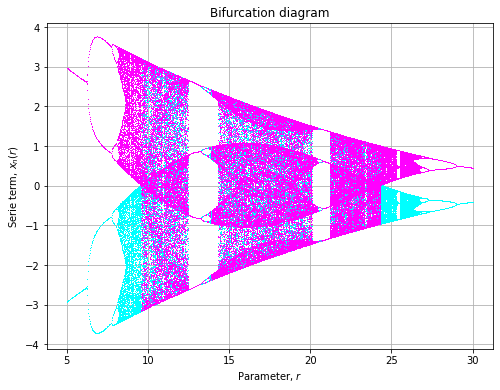
)Sinusoidal
![]()
def sinusuodal(x, r, p=0.7):
return r * x ** 2 * np.sin(np.pi * x) + p
axe = plot(
model=sinusuodal,
rmin=-2.7, rmax=1.4,
xlin=np.array([-0.1, 0.1])
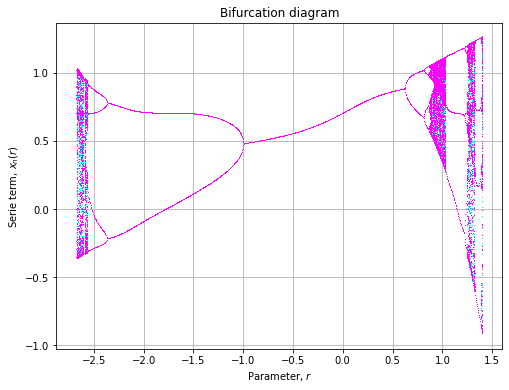
)Cosinusoidal
![]()
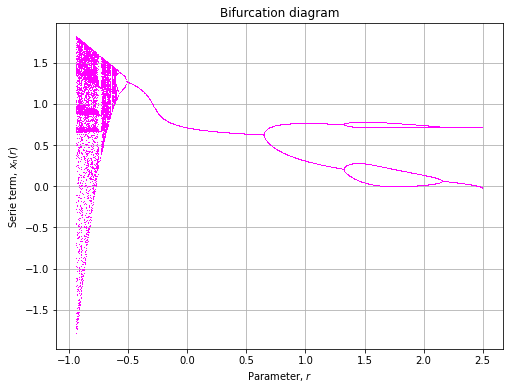
def cosinusuodal(x, r, p=0.7):
return r * x ** 2 * np.cos(np.pi * x) + p
axe = plot(
model=cosinusuodal,
rmin=-1, rmax=2.5,
xlin=np.array([-0.1, 0.1])
)