In this post we show how an integral can be computed using Monte-Carlo method and how combination with bootstrapping can increase precision.
Reference
Lets say we wan to integrate the cardinal sinus over the interval ![]() . Graphically it is about finding the area under the curve by taking care to respect sign of positive (above x axis) and negative area (below x axis).
. Graphically it is about finding the area under the curve by taking care to respect sign of positive (above x axis) and negative area (below x axis).
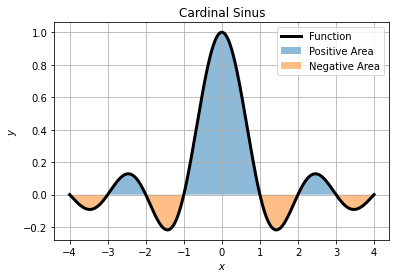
The area of this curve can be numerically computed by trapezoid or quadrature, or analytically.
def model(x):
return np.sinc(x)
xlin = np.linspace(-4., 4., 20000)
ylin = model(xlin)
Itrap = integrate.trapezoid(ylin, x=xlin)
# 0.9499393464346436
Iquad = integrate.quad(model, xlin.min(), xlin.max())
# (0.9499393397673102, 9.485684460130983e-11)Both method agrees with the 6 first significant figures and the quadrature value agrees up to its precision to the approximation given by Wolfram Alpha:
![]()
Method
First we must state the transfer model:
![Rendered by QuickLaTeX.com \[I = \int\limits_a^b f(x) \mathrm{d}x\]](https://landercy.be/wp-content/ql-cache/quicklatex.com-cd2faf22a7717ae761a739822d1629bf_l3.png)
Which can be approximated by:
![Rendered by QuickLaTeX.com \[I \simeq \frac{b - a}{N} \sum\limits_{i=1}^N f(a + (b-a) \cdot U_{0,1})\]](https://landercy.be/wp-content/ql-cache/quicklatex.com-805e3b850b7c1bf3bc2cc42ecfe02b87_l3.png)
The Monte-Carlo method works as follow:
- Uniformly randomly sample
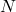 points on the domain extent such as
points on the domain extent such as 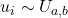 ;
; - Compute the image of all this points
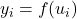 ;
; - Estimate the integral
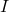 using the approximation of the transfer model ;
using the approximation of the transfer model ; - Repeat the process
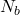 time to bootstrap the procedure and let the CLT come into action;
time to bootstrap the procedure and let the CLT come into action;
The function below implements such a procedure except bootstrapping:
def monte_carlo(N, a=-1., b=1., model=lambda x: x, seed=12345):
np.random.seed(seed)
x = np.random.uniform(a, b, size=N)
y = model(x)
return (b - a) / N * np.sum(y)A single trial gives a poor estimate of the integral, hence the need of bootstrapping:
I = monte_carlo(100000, a=-4., b=4., model=model)
# 0.9491438852706744Now we will repeat the procedure for different sample size ![]() and for each population size we will bootstrap to enhance the estimation:
and for each population size we will bootstrap to enhance the estimation:
Nb = 3000
np.random.seed(12345)
estimates = []
for s in np.random.randint(0, 999999999, size=Nb):
for n in np.logspace(4, 7, 30, base=10).astype(int):
I = monte_carlo(n, a=-4., b=4., model=model, seed=s)
estimates.append({"s": s, "n": n, "I": I})
data = pd.DataFrame(estimates)Once the bootstrap finishes we can aggregate bootstrap samples to see how it helps to converge towards the expected integral value:
agg = data.groupby("n")["I"].agg(["mean", "std"])Graphically it leads to:
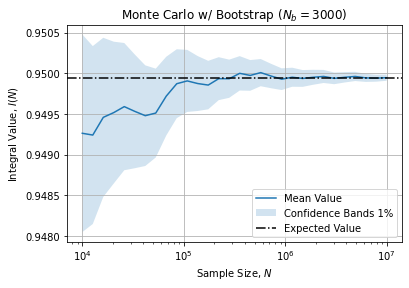
Which shows a convergence towards the expected value when sample ![]() increases. Additionally bootstrap samples allows to compute Confidence Bands (here 1%).
increases. Additionally bootstrap samples allows to compute Confidence Bands (here 1%).
