In this post we propose a simple implementation of Q-Q Plot and show some basic usage of it.
Implementation
A clean definition of a Q-Q Plot is:
In statistics, a Q–Q plot (quantile–quantile plot) is a probability plot, a graphical method for comparing two probability distributions by plotting their quantiles against each other
Wikipedia
Usually Q-Q plot compares a theoretical distribution/law with an experimental dataset. The procedure is then:
- Compute the ECDF of dataset to get empirical quantiles;
- Chose a theoretical model, you may fit against your data (eg. using MLE);
- Compute theoretical quantiles using quantile function (or PPF) from you theoretical model;
- Create pair of quantiles (theoretical, empirical).
If the Q-Q points lie on the ![]() line, distribution perfectly agrees. If there are deviation it informs in which direction, so we can infer the behaviour.
line, distribution perfectly agrees. If there are deviation it informs in which direction, so we can infer the behaviour.
The following snippet implement such a procedure:
def qqplot(data, law_factory, axe=None):
if axe is None:
fig, axe = plt.subplots()
# Compute ECDF from data:
ecdf = stats.ecdf(data)
# Check if law is already parametered:
if isinstance(law_factory, stats._distn_infrastructure.rv_continuous_frozen):
law = law_factory
# Fit using MLE if not the case:
else:
parameters = law_factory.fit(data)
law = law_factory(*parameters)
# Compute theoretical quantiles:
quantiles = law.ppf(ecdf.cdf.probabilities)
axe.scatter(quantiles, ecdf.cdf.quantiles, marker=".")
axe.plot(quantiles, quantiles, "--", color="black")
axe.set_title("Q-Q Plot: %s\n args=%s, kwargs=%s" % (law.dist.name, np.array(law.args), law.kwds))
axe.set_xlabel("Theoretical Quantile")
axe.set_ylabel("Empirical Quantile")
axe.grid()
return axe, lawThe function can receive either a generic random variable or a parameterized one.
Usage
Lets draw some sample from a Chi Square distribution:
np.random.seed(12345)
law = stats.chi2(df=30)
data = law.rvs(size=30_000)Now lets compare it with respect to a Gaussian:
axe, law1 = qqplot(data, stats.norm)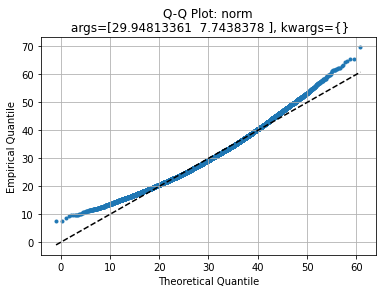
We see that central values agrees with theoretical quantiles but left and right tails deviate.
If we compare it with a generic Chi Square distribution:
axe, law2 = qqplot(data, stats.chi2)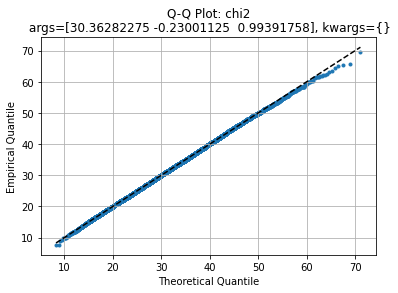
We get an almost straight line, indicating the model may suit for this dataset. Comparing with the original distribution:
axe, law2 = qqplot(data, law)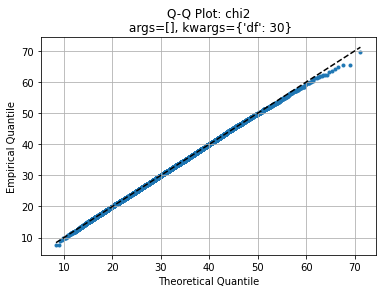
The agreement is identical. Challenging against a Log Normal we have:
axe, law2 = qqplot(data, stats.lognorm)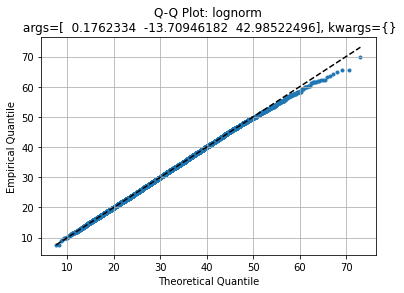
Which quite agrees as well but with a slightly fatter right tail.
Comparison
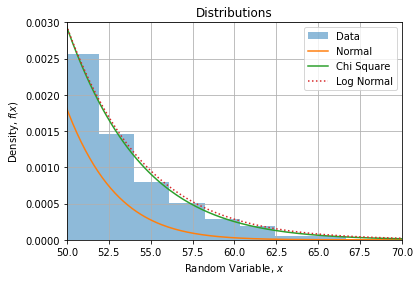
If we compare CDF’s with data histogram, we find the same observations:
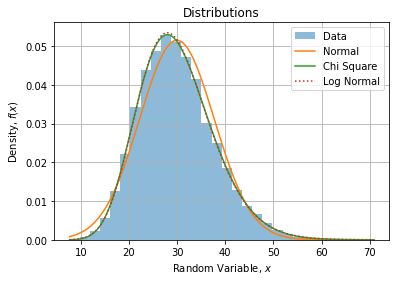
If we zoom on the right tail, we can confirm Log Normal is a bit fatter than Chi Square: